Nuclear and Many Body Physics
Our group works on mathematical and computational models of quantum many-body systems, mostly nuclei but also the electronic structure of atoms and gases of cold atoms.
Some of the topics within this area include:
- High performance computation of nuclear structure,
- First principles calculations of nuclear structure,
- Exploring how orderly patterns arise out of symmetry and randomness,
- Scattering and reactions in nuclei and in atomic systems,
- Calculation of weak transitions in nuclei for astrophysics, including electron capture and interactions with neutrinos.
Our major tool is diagonalizing the many-body Hamiltonian, a difficult problem which requires sophisticated algorithms implemented on parallel supercomputer. Towards this end we use Fortran 90/95 and MPI and OpenMP parallelization tools. We also develop and benchmark alternate approaches to solving the many-body Hamiltonian.
The research is supported by the U.S. Department of Energy and the best students often spend summers at national labs, such as Lawrence Livermore, Los Alamos, and Oak Ridge National Laboratories. Two of our past students have permanent staff positions at national labs.
Students who are interested in fundamental physics and have an interest in cutting-edge, hands-on computing will enjoy this research.
For more information, contact Professor Calvin Johnson.
Polymer & Biological Physics
Dr. Baljon was a faculty at SDSU from 1999-2025. Currently she is still working half time. Students interested in a special study or research project in the area of polymer/biological physics can contact her by emailing abaljon@sdsu.edu.
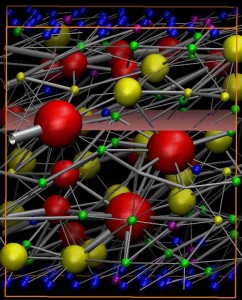
Dr. Baljon’s research is on associating polymers. In a novel MD/MC simulation the polymers are modeled as strings of beads, the ends of which can connect to each other. These connections are temporary and can break due to temporal fluctuations or applied stress. New connections form when polymers come close to each other. At high concentration and low temperature a network forms (see picture), the topology of which can be characterized using network theory. She has published on the changes in network topology that occur when the system is under a uniform or oscillatory stress.
In the area of biophysics, she studied the morphology of the inner mitochondrial membrane in collaboration with faculty in biology (Dr. Frey) and mathematics (Dr. Salamon and Dr. Nulton).
More recently she models mucus. Biologist have shown that mucus helps phage to protect the body against bacterial pathogens and is crucial for a healthy microbiome. As any adaptive polymeric network, mucus encodes information about its environment into its structural and dynamical patterns. How does mucus help phage to hunt for bacteria and defend the human body? Is healthy mucus wired or rewiring in a specific way?
Both undergraduate and graduate students are actively involved in this research and most are coauthors of professional publications. Many have attended conferences and presented posters or short talks. UG can join the group during the semester in which they take junior level classical mechanics. Graduate students are invited to join during their first semester at SDSU. Currently summer stipends are available for 2014 and 2015. Students who are already involved in the research are given preference over new students. If you are interested, please stop by my office (room P-134).